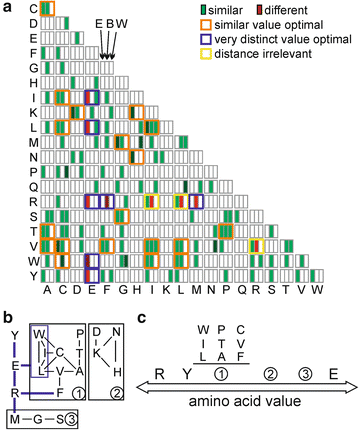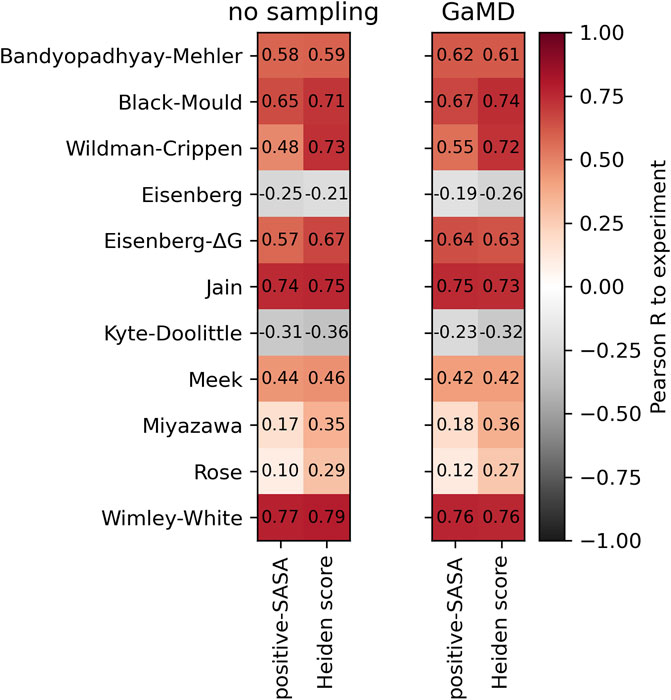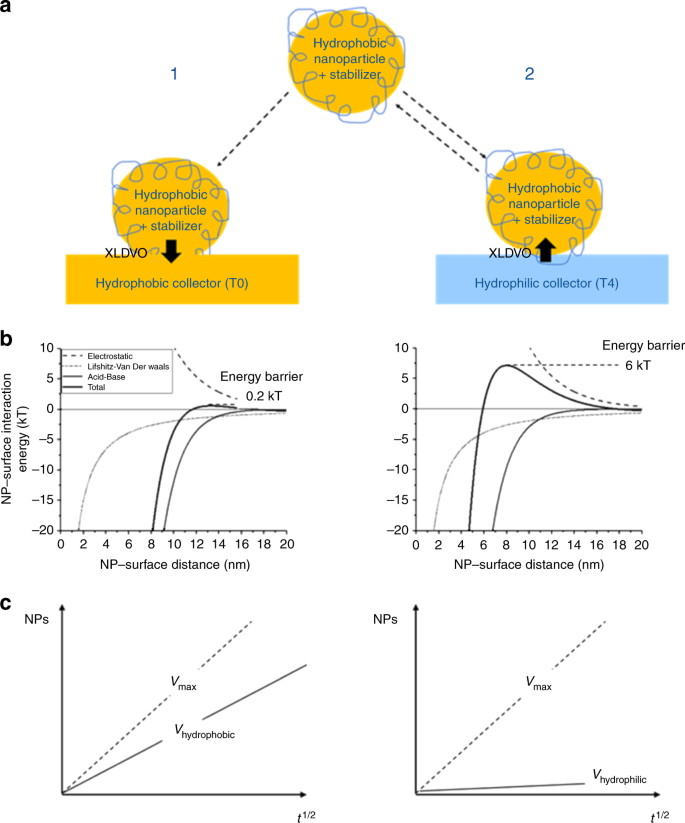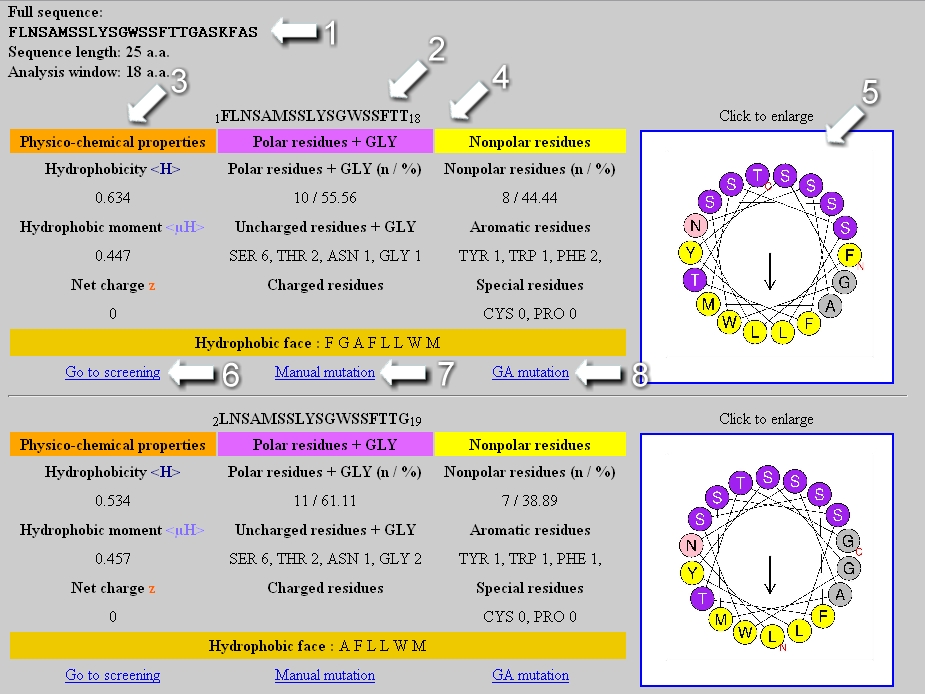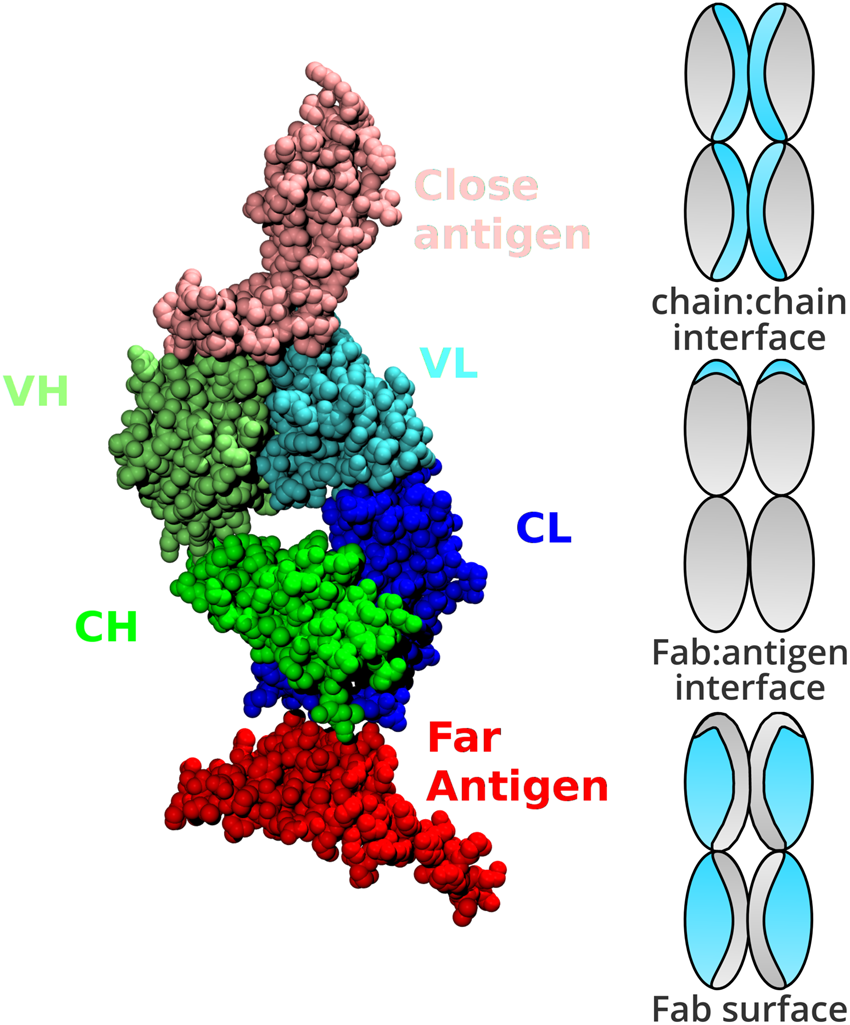
Web-based display of protein surface and pH-dependent properties for assessing the developability of biotherapeutics | Scientific Reports
RNase k-02 hydrophobicity plot and Western blot analysis of HEK-293... | Download Scientific Diagram

Equation 1 was used to determine effective hydrophobicity. The helical... | Download Scientific Diagram

100th Anniversary of Macromolecular Science Viewpoint: The Role of Hydrophobicity in Polymer Phenomena | ACS Macro Letters

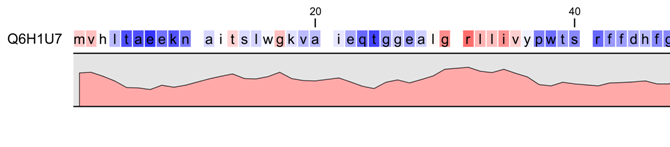
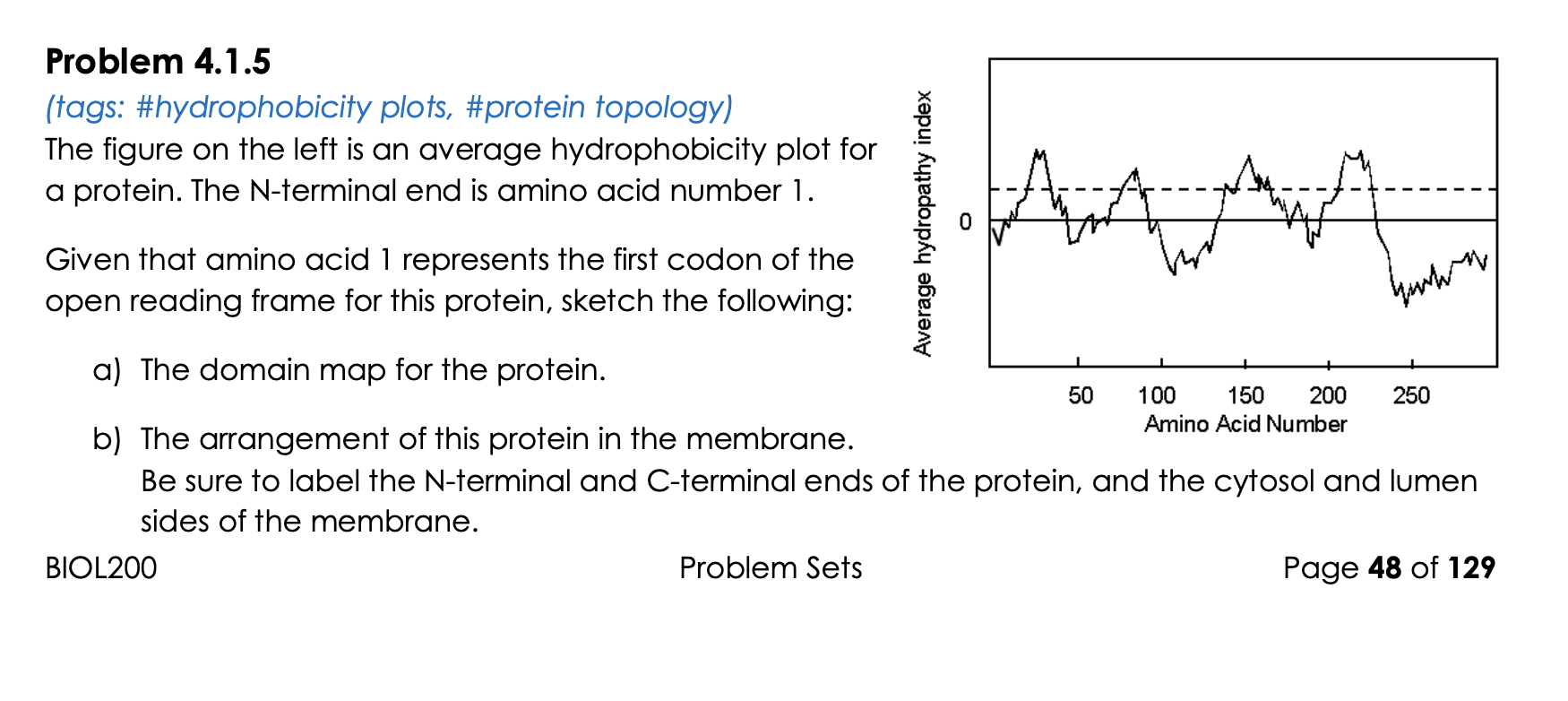
SOLVED: The complete sequence of Band 3 is available: We analyze its amino acid primary sequence to obtain a hydrophobicity plot represented here: ProtScale output for user sequence Hphob Kyte Doolittle 100

Methods of calculating protein hydrophobicity and their application in developing correlations to predict hydrophobic interaction chromatography retention - ScienceDirect

An Intrinsic Hydrophobicity Scale for Amino Acids and Its Application to Fluorinated Compounds - Hoffmann - 2019 - Angewandte Chemie International Edition - Wiley Online Library